Getting started¶
If you have a supported Python installation on your computer, you can install the package via
pip install scikit-fem[all]
Specifying [all] includes meshio for mesh input/output, and
matplotlib for simple visualizations. The minimal dependencies are
numpy and scipy. You can also install scikit-fem in Google Colab by executing
!pip install scikit-fem
Step 1: Clarify the problem¶
In this tutorial we solve the Poisson problem
where \(\Omega = (0, 1)^2\) is a square domain and \(f(x,y)=\sin \pi x \sin \pi y\). The weak formulation reads: find \(u \in H^1_0(\Omega)\) satisfying
Note
\(H^1_0(\Omega)\) is the space of functions that are zero on the boundary \(\partial \Omega\) and have finite energy: the square integral of the first derivative is finite.
Step 2: Express the forms as code¶
Next we write the forms
as source code. Each form is written as a function and decorated as follows:
>>> import skfem as fem
>>> from skfem.helpers import dot, grad # helpers make forms look nice
>>> @fem.BilinearForm
... def a(u, v, _):
... return dot(grad(u), grad(v))
>>> import numpy as np
>>> @fem.LinearForm
... def L(v, w):
... x, y = w.x # global coordinates
... f = np.sin(np.pi * x) * np.sin(np.pi * y)
... return f * v
For more information see Anatomy of forms.
Step 3: Create a mesh¶
The default constructors of Mesh initialize a
unit square:
>>> mesh = fem.MeshTri().refined(3) # refine thrice
>>> mesh
<skfem MeshTri1 object>
Number of elements: 128
Number of vertices: 81
Number of nodes: 81
Named boundaries [# facets]: left [8], bottom [8], right [8], top [8]
Step 4: Define a basis¶
The mesh is combined with a finite element to form a global basis. Here we choose the piecewise-linear basis:
>>> Vh = fem.Basis(mesh, fem.ElementTriP1())
>>> Vh
<skfem CellBasis(MeshTri1, ElementTriP1) object>
Number of elements: 128
Number of DOFs: 81
Size: 27648 B
Step 5: Assemble the linear system¶
Now everything is in place for the finite element assembly.
The resulting matrix has the type scipy.sparse.csr_matrix
and the load vector has the type ndarray.
>>> A = a.assemble(Vh)
>>> l = L.assemble(Vh)
>>> A.shape
(81, 81)
>>> l.shape
(81,)
Step 6: Find boundary DOFs¶
Setting boundary conditions requires finding the degrees-of-freedom (DOFs) on
the boundary. Empty call to
get_dofs() matches all boundary DOFs.
>>> D = Vh.get_dofs()
>>> D
<skfem DofsView(MeshTri1, ElementTriP1) object>
Number of nodal DOFs: 32 ['u']
Step 7: Eliminate boundary DOFs and solve¶
The boundary DOFs must be eliminated from the linear system \(Ax=l\)
to set \(u=0\) on the boundary.
This can be done using condense().
The output can be passed to solve()
which is a simple wrapper to scipy sparse solver:
>>> x = fem.solve(*fem.condense(A, l, D=D))
>>> x.shape
(81,)
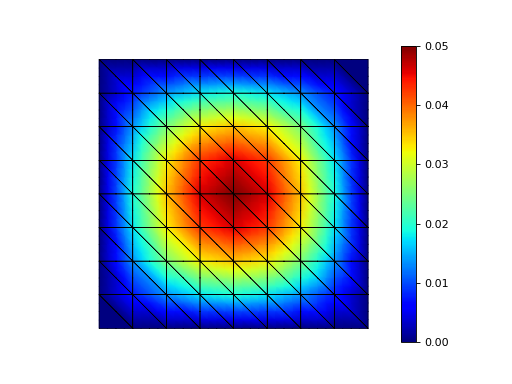
Step 8: Calculate error¶
The exact solution is known to be
Thus, it makes sense to verify that the error is small.
>>> @fem.Functional
... def error(w):
... x, y = w.x
... uh = w['uh']
... u = np.sin(np.pi * x) * np.sin(np.pi * y) / (2. * np.pi ** 2)
... return (uh - u) ** 2
>>> str(round(error.assemble(Vh, uh=Vh.interpolate(x)), 9))
'1.069e-06'